J. Northw. Atl. Fish. Sci., Vol. 53: 35–45
Publication (Upload) date: 08 Sep. 2022
Robert D. Murphy Jr.1,2* , Gary A. Nelson3
, Gary A. Nelson3 ,
, Jonathan H. Grabowski1
Jonathan H. Grabowski1

1Department of Marine and Environmental Sciences, Marine Science Center, Northeastern University,
430 Nahant Road, Nahant, MA 01908, USA
2Alaska Pacific University, Fisheries, Aquatic Science and Technology Laboratory, 4101
University Drive, Anchorage, AK 99508, USA
3Massachusetts Division of Marine Fisheries, Cat Cove Laboratory, 92 Fort Avenue,
Salem, MA 01970, USA
*E-mail: rdmurphy@alaskapacific.edu (RM)
Murphy Jr., R.D., Nelson, G.A., and Grabowski, J.H. 2022. The feeding ecology of striped bass and the role of ontogeny. J. Northw. Atl. Fish. Sci., 53: 35–45. https://doi.org/10.2960/J.v53.m737
Abstract
Amidst constantly changing biotic and abiotic conditions, a more thorough understanding of the ecological consequences of dynamic predator-prey interactions will likely enable increasingly sustainable fisheries management. This study assessed the diet of striped bass, a generalist marine predator in coastal Massachusetts that feed on a variety of prey species and impose top-down pressure on other important fishery species, such as the American lobster and Atlantic menhaden. We explored the role of ontogeny using both stomach content and stable isotope analyses. Empirical results from 158 striped bass collected in northern Massachusetts revealed that striped bass in this area may have shifted from feeding predominantly on Atlantic menhaden in the late 1990s and early 2000s to Atlantic mackerel in this study. Stable isotope data suggested that the diet of striped bass is significantly linked with ontogeny: larger fish feed more heavily on benthic prey, particularly in the latter half of their seasonal residency in Massachusetts. Our study suggests that large striped bass gain an energetic advantage, as indicated by a liver somatic index, by feeding on benthic prey, possibly due to decreased foraging costs. Collectively, this work illustrates the ability of predatory fish to capitalize on the variability of forage fish populations, but highlights the importance of invertebrate prey for large striped bass and proposes underlying mechanisms driving ontogenetic diet switches from piscivory to benthivory.
Keywords:striped bass (Morone saxatilis), ontogenic diet switch, predator-prey interactions, stomach content analysis, stable isotope analysis
PDF | Supp. Materials.
Download Citation Data

Citation to clipboard
 Reference management software (Endnote, Mendeley, RefWords, Zotero & most other reference management software)
Reference management software (Endnote, Mendeley, RefWords, Zotero & most other reference management software)
LaTex, BibDesk & other specific software
Introduction
Comprehensive knowledge of predator-prey interactions is an important underpinning of ecosystem approaches to fisheries management, which is increasingly receiving attention (e.g., North Pacific Fishery Management Council 2019). Further, temporal variability in species distributions, abundances, and size-frequencies necessitates that we routinely monitor ecosystems and the consequences of changing predator-prey interactions (Hilborn et al., 2017). For example, predators can have strong top-down effects on prey populations and alter ecosystems (Denno and Lewis, 2009). They exert control over the distribution of species (Connell, 1961), mediate trophic cascades (Carpenter et al., 1985), and control the flow of nutrients within food webs (Trussell et al., 2006; Hawlena and Schmitz, 2010). Conversely, the availability of prey can fundamentally affect predators (Sherwood et al., 2002). For instance, along the coast of Canada, declining capelin (Mallotus villosus) availability (an important prey species) may have contributed to reduced lipid storage and spawning potential in Atlantic cod (Gadus morhua) (Sherwood et al., 2007). There is growing evidence, however, that the reliance of a predator population on forage fish abundance is largely context-dependent. Hilborn and colleagues (2017) argued that forage fish abundance rarely has a measurable impact on predator abundance in U.S. fisheries, in part because predators may exhibit significant behavioral plasticity and modify their feeding to account for the natural variability of prey populations.
An important step in investigating predator-prey interactions involves characterizing the suite of factors that can alter prey selection (Juanes et al., 1994). Optimal Foraging Theory (OFT) suggests that a predator will select prey items by balancing the costs of energy acquisition and consumption relative to the intake of energy. More specifically, OFT predicts that predators will select prey that maximize the difference between the energetic value of the prey and the energetic cost of pursuing, attacking, and handling the prey (Pyke et al., 1977). However, other factors may prevent predators from consuming optimal prey, including the presence of competitors (intra- and inter-specific competition), avoidance of their own predators, and morphological limitations such as gape width (Hughes, 1990; Hambright, 1991; Einfalt and Wahl, 1997). For instance, Milinksi (1982) found that sticklebacks consumed fewer optimal prey items in the presence of superior intra-specific competitors. Additionally, optimal prey may change as predators grow, as they may “switch” to consuming a completely new, often larger prey type (i.e., an ontogenetic diet shift) to overcome the aerobic and anaerobic costs of prey consumption (Townsend and Winfield, 1985; Sherwood et al., 2002). Fluctuations in the abundance of prey populations may, however, drive predators to consume less energy-dense but more abundant prey, leading to declines in predator condition (Sherwood et al., 2007).
In the western Atlantic, significant historic and more recent fluctuations in the abundance of both striped bass (Morone saxatilis) and their prey emphasize that predator-prey interactions are dynamic (Hill et al., 1989; Atlantic States Marine Fisheries Commission, 2014). The striped bass is a highly mobile and generalist predator that typically spawns in western Atlantic, mid-coast United States estuaries and brackish habitats and migrates north during the spring and summer where they feed heavily on economically valuable prey species like the American lobster (Homarus americanus) and Atlantic menhaden (Brevoortia tyrannus) (Bigelow et al., 1953, Boreman et al., 1987; Nelson et al., 2003). However, striped bass also consumed large quantities of the Blueback herring (Alosa aestivalis) (Greene et al., 2009), which has since declined in many coastal and riverine ecosystems (Atlantic States Marine Fisheries Commission, 2017). Striped bass may have shifted their diet in the late 1990s towards other Clupeid prey that were still prevalent like the Atlantic menhaden (Nelson et al., 2003). Our ability to sustainably manage both striped bass and their prey populations amidst such dynamic ecosystems will require a holistic understanding of the causes and consequences of these interactions across space and time.
Our study explored the feeding ecology and potential role of ontogeny in striped bass during their spring and summer migration into Massachusetts (MA) where they consume a variety of prey items from zooplankton and fish to large invertebrates, such as the American lobster and green crab (Carcinus maenas) (Chapoton and Sykes, 1961; Manooch, 1973; Nelson et al., 2003). Striped bass spawn and spend the majority of the year in the western Atlantic, mid-coast United States, and the vast majority of studies on striped bass feeding ecology have been conducted in the southern half of their range, such as the Chesapeake Bay (Dovel, 1968; Gardinier and Hoff, 1982; Dunning et al., 1997; Griffin and Margraf, 2003). By and large, these studies indicate that juveniles feed on zooplankton and small crustaceans, while adult striped bass are predominately piscivorous, but may also consume a small proportion of invertebrate prey (Manooch, 1973; Gardinier and Hoff, 1982; Griffin and Margraf, 2003; Overton et al., 2009). In contrast to these studies, Nelson et al. (2003, 2006) conducted an extensive diet study on striped bass collected between 1997–2000, whereby half of the collected fish were from the North Shore region of coastal MA. Their results suggested that as striped bass grow, they rely more heavily on benthic decapod prey, while smaller adults feed more on forage fish. This apparent ontogenetic diet shift may have other consequences on striped bass since crustaceans, such as American lobsters, may generate proportionally less energy per gram wet weight as compared to forage fish such as Atlantic herring, Clupea harrengus (Nelson et al., 2006). Crustaceans also require more time for predatory fish to digest them (Langton and Center, 1982) and, as such, may represent a suboptimal prey choice. The mechanisms for this potential ontogenetic prey shift are unclear, along with the degree to which suboptimal prey consumption influences the condition and growth of striped bass (Sherwood et al., 2002).
To assess the diet of striped bass in northern MA, we conducted stomach content and stable isotope analyses. Traditional stomach content analysis can result in precise identification of prey species but offers only a recent snapshot of what an individual has been consuming. An alternative approach, utilizing stable isotopic ratios in predator tissue, provides an approximate yet more holistic metric because it examines the assimilation of consumed prey into predator biomass. The stable isotope ratios of nitrogen (δ14N / δ15N) indicate trophic position due to the predictable enrichment of nitrogen for predators relative to their prey (Fry, 1988; Post, 2002). Conversely, the stable isotope ratios of carbon (δ13C / δ12C) do not fractionate as much between trophic levels, and thus indicate benthic versus pelagic feeding due to differences in the enrichment of carbon isotopes at the base of the food chain (Post, 2002). As such, our study used stomach content and stable isotope analyses to identify important prey taxa, evaluate the role of ontogeny, and explore whether diet metrics correlate with predator condition. Thus, we used this approach to explore possible mechanisms underlying diet transitions in striped bass.
Materials and Methods
All methods were approved by Northeastern University’s Institutional Animal Care and Use Committee. From 2012 to 2016, striped bass were collected via rod-and-reel from the North Shore region of MA between Nahant and Gloucester, centralized around Salem Sound (Fig. 1; n = 158, total length (TL) range = 41.3cm–111.8cm, mean = 77.4cm). Once caught, fish were euthanized via pithing, and TL and fork length of a fish were measured to the nearest tenth of a cm, and fish were placed on ice. A small white muscle plug was extracted from an area 1–3cm below the first dorsal fin for stable isotope analysis and was immediately placed in foil and frozen.
Fig. 1
In the laboratory, stomach contents were extracted, and prey items were identified to the lowest taxon possible. The number of individuals by species and the weight (g) of each species were recorded. Prey specimens in good condition (i.e., not digested) were saved and frozen for stable isotope analysis. Samples for stable isotope analysis were taken internally from prey as to reduce the likelihood of contamination. Multiple metrics were used to examine the importance of prey taxon for striped bass. First, empty stomachs were removed from further stomach content analysis (these fish were included in stable isotope analyses, however). Percent weight (W) is a useful metric for comparing the relative energetic value of prey, especially when individuals from different taxa are of disparate sizes (Zale et al., 2012). Percent weight was calculated as the fraction of the total weight of an individual taxon by the total weight of stomach contents for all fish with non-empty stomachs. To determine how often striped bass consumed particular prey, we calculated the frequency of occurrence (F) for each prey item: the fraction of stomachs with an individual taxon by the total number of non-empty stomachs. Both W and F metrics were determined for all striped bass and by size category, whereby fish with non-empty stomachs were separated based on those below and above the mean TL (mean TL was calculated based on non-empty stomachs). To examine the importance of forage fish versus benthic decapods, the following prey items were classified as benthic decapods: Jonah crabs (Cancer borealis), rock crabs, green crabs, unclassified crabs (Cancridae), unclassified decapods (Decapod), Asian shore crabs (Hemigrapsus sanguineus), and American lobsters. While sand shrimp (Crangon septemspinosa) is of the Order Decapoda, we did not classify it as a benthic decapod given its propensity to swim off the substrate, and the different striped bass attack strategies used to consume Sand shrimp versus other, larger benthic decapods (note, only two Sand shrimp were found in striped bass stomachs in our study).
To further explore the role of ontogeny in diet, stable isotope analysis was used as a longer-term approximation of predator diet because it measures prey that have been assimilated into muscle and other tissues (Post, 2002). Using sterile techniques, a small internal plug from each frozen sample was collected internally to avoid contamination. Each sample was then dried for 48 hours at 45°C and subsequently ground to a homogenous powder using a sterilized mortar and pestle. Samples were then weighed, placed in tin caps, and packed for shipment. All samples, including 10% duplicates (i.e., separate sub-samples were taken from the same tissue to examine variation between replicate pairs), were sent to the Colorado Plateau Laboratories to be analyzed. Since lipid content can influence isotopic carbon signatures, prey and predator samples (Post et al., 2007) were lipid-corrected by using methods of Skinner et al. (2016). As such, lipid percentages were generated based on a formula from Post et al. (2007), which was used as an input in another formula from Kiljunen et al. (2006) to correct δ13C values (hereby called δ13C’).
To estimate predator condition, two metrics were utilized. Liver somatic index (LSI) was calculated for each fish as: LSI = wet liver weight / wet body weight * 100 (Adams and McLean, 1985). An individual fish’s LSI should be closely correlated with health since excess energy is stored as glycogen in the liver, typically after periods of high prey consumption (Hoque et al., 1998). Thus, higher relative LSI values should indicate a healthier individual with greater energy stores. To explore the effects of diet on striped bass relative body size, the Relative Condition Factor (Kn) was used, which is standardized to account for allometric growth (Le Cren, 1951). Here, individual fish weight (W) was divided by the length specific mean weight (w’) of striped bass in MA such that K = w / w’. Length specific mean weight was calculated according to the MA striped bass Monitoring Report for 2014: log10 (Wp) = -3.455 + 3.001 * log10(Li, where Wp is weight in pounds (1 pound = 454 grams) and Li is the total length in inches (1 inch = 2.54 cm) (Nelson, 2015).
Statistical Analysis
Linear regression models were used to explore the potential relationships between striped bass stable isotopic values for δ13C’ and δ15N and striped bass TL and the day of year (day). We also examined the relationships between response variables, LSI and Kn, and potential predictors, striped bass TL and δ13C’, as a proxy for benthic feeding using linear regression models. As such, the four models were as follows; (1) δ13C’ = f (striped bass TL * day);(2) δ15N = f (striped bass TL * day);(3) LSI = f (striped bass TL * δ13C’); (4) Kn = f (striped bass TL * δ13C’). All data were modeled as normally-distributed errors (Gaussian GLM) in R (R Core Team, 2020) and models included an interactive term between covariate predictors. Assumptions of residual normality were assessed using normal quantiles plots, while homoscedasticity was inspected using residuals versus fitted values plots. Regression terms were deemed significant at α ≤ 0.05.
Results
Atlantic mackerel, Scomber scrombrus, was the most important prey item by weight (42.8%), followed by American lobster (19.9%), unclassified fish prey (10.3%), and Menhaden (7.0%), while several other species were of much lower importance (Fig. 2). Frequency of occurrence of unclassified fish prey (33.3%), Atlantic mackerel (17.7%), American lobster (16.7%), and rock crabs, Cancer irroratus, (16.7%) were higher than other prey taxa consumed by striped bass (Table 1, Fig. 2a). When aggregated by size, small striped bass (i.e., those below the 76.54cm mean TL for non-empty fish) and large striped bass had consumed a similar amount of fish prey, representing 79.5% and 61.2% of their diet by weight, and were found in 58.8% and 64.7% of non-empty stomachs, respectively (Fig. 2b). Conversely, large striped bass consumed more benthic decapods by weight (35.5%) and more frequently (51%) compared to small striped bass (13.2% and 35.3%, respectively). Examination of all striped bass (i.e., those with prey in their stomachs and those with empty stomachs) revealed that the large and small fish had empty stomachs 39% and 32% of the time, respectively.

Fig. 2
Stable isotope analysis revealed little variation between replicate pairs (mean of absolute differences between pairs) for striped bass muscle samples (δ13C’ = 0.19‰, δ15N = 0.20‰, n = 15). Striped bass stable isotopic values were adjusted to account for trophic fractionation between predator and prey (δ13C’ = +0.8‰, δ15N = +3.4‰, Fig. 3a) and then for plotting purposes and to visually compare striped bass to their prey, mean isotopic values for striped bass were plotted alongside prey values (Fig. 3b) (Zanden and Rasmussen, 2001). Both species of prey fish, Atlantic herring and Atlantic mackerel, had the highest δ15N and lowest δ13C’ values among prey, indicating that they represent a higher trophic level and consume a more pelagic food source than crustacean prey. Meanwhile, the two crab species, green crab and rock crab, and the American lobster were of a lower trophic level and δ13C’ was highly enriched, indicating that these species represent benthic prey consumed by striped bass (Fig. 3a).
A model of stable carbon isotopes revealed a significant interaction between TL and day (degrees of freedom (DF) = 151, t-value = 2.456, p-value = 0.015; Fig. 4a). To aid in the visualization of this significant interaction, we conducted linear regression and quantified the slopes for the relationship between δ13C’ and day for striped bass whose length was ±1 standard deviation from the mean, which revealed a positive slope for larger fish and negative slope for smaller fish (Fig. 4b). TL and day were not significantly related to δ15N (DF = 151, TL: t = -1.173, p = 0.243; day: t = -1.863, p = 0.064; TL x day: t = 1.569, p = 0.119).
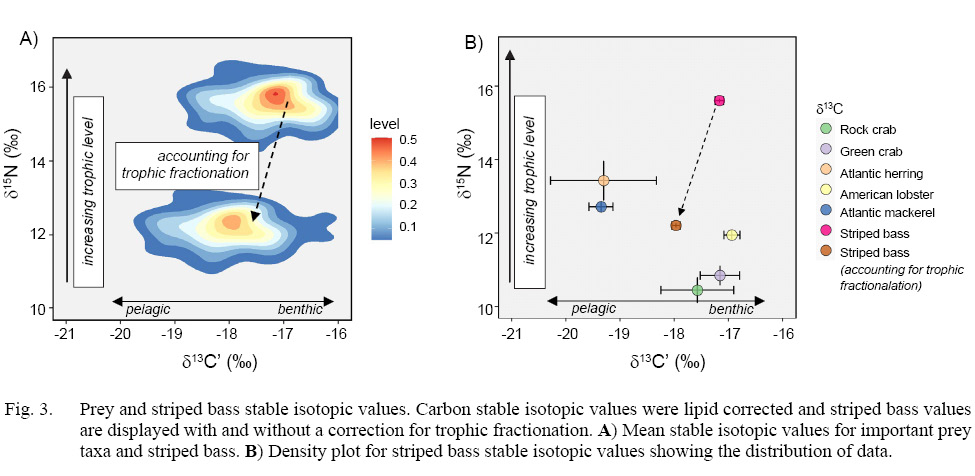
Fig. 3
Examination of condition indices revealed significant interactions between stable carbon isotopic values, TL, and LSI (DF = 147, t = 2.593, p = 0.011; Fig. 5a). Again, we used linear regression to quantify the slopes for the relationship between LSI and δ13C’ for striped bass whose length was ±1 standard deviation from the mean to aid in result interpretation. This revealed a positive slope for larger fish and negative slope for smaller fish (Fig. 5b). Lastly, Kn was not significantly related to δ13C’ and TL (DF = 148, δ13C’: t = -1.731, p = 0.09; TL: t = 1.798, p = 0.074; TL x δ13C’: t = 1.707, p = 0.09).
Discussion
Our results indicated that striped bass in the North Shore region of MA maintain a diet high in Atlantic mackerel, which is in contrast to research two decades earlier that suggested striped bass diets were dominated by Atlantic menhaden (Nelson et al., 2003). The occurrence of Atlantic mackerel in striped bass diets increased over 10-fold, potentially indicating a major shift in local availability of forage fish. As opportunistic predators, striped bass appeared to have capitalized on the local variability of forage fish populations (Hilborn et al., 2017). Additionally, Nelson et al. (2003) found that striped bass similar in size to those sampled in this study consumed predominantly Atlantic menhaden in the later summer months, which is when this forage fish typically migrates into nearshore waters along coastal MA. Anecdotal evidence suggests that Atlantic menhaden abundances in Salem Sound have been very low in recent years, whereas there was a considerable uptick in Atlantic mackerel spawning-stock-biomass and total biomass directly following the Nelson et al. (2003) diet study (42nd Northeast Regional Stock Assessment Workshop, 2006).
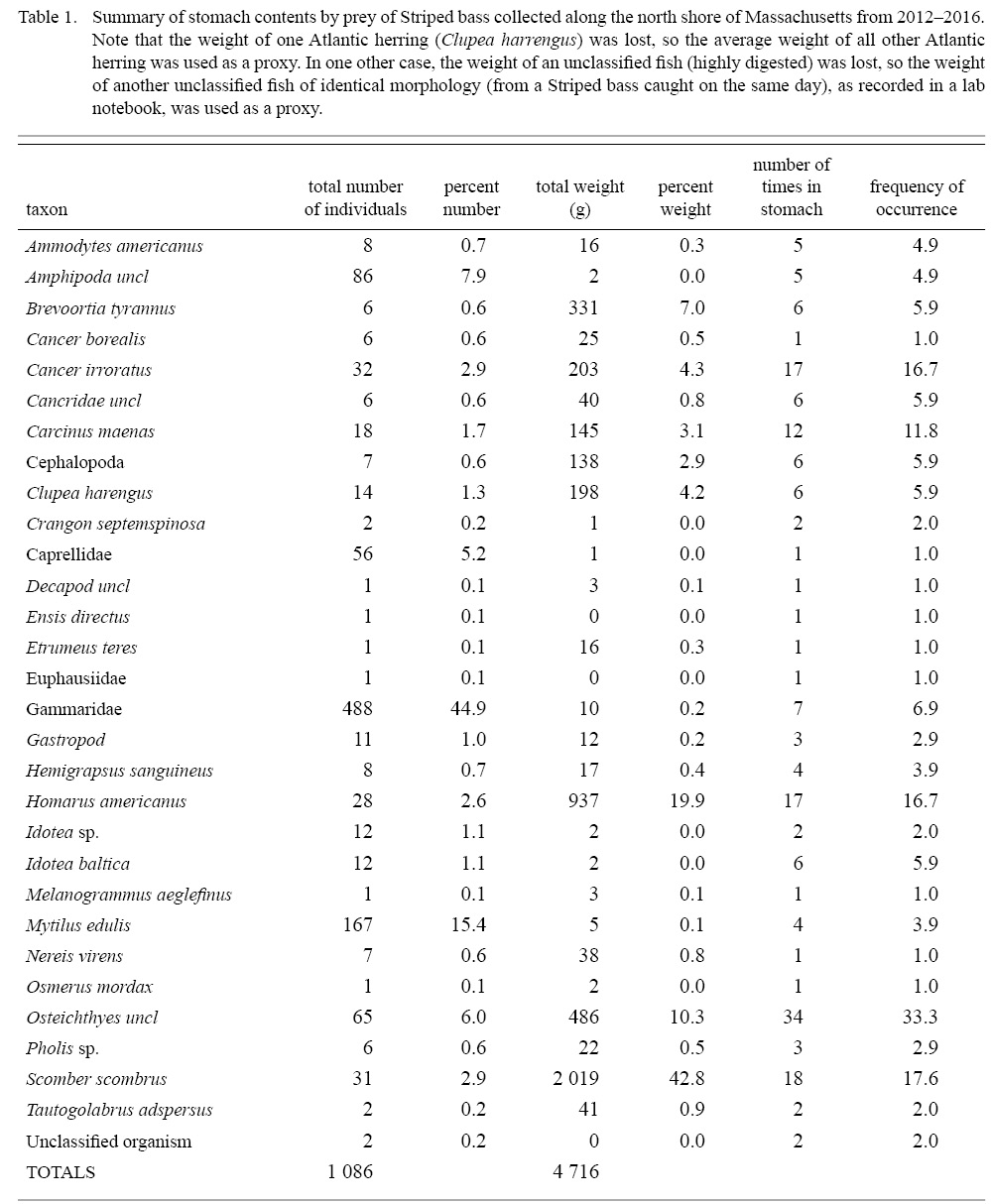
Table 1
Stomach content analysis revealed that the American lobster may also be a critical prey item, and was the most important invertebrate taxa by weight, highlighting an interaction with another vital New England fishery. Catch of American lobster in MA was valued at over $82 million in 2016, second only to Sea scallops (MA Division of Marine Fisheries Annual Report, 2016). Rock crabs were consumed at similar rates but are much smaller and thus likely represent a lesser energy source. This finding agrees with Nelson et al. (2003) in their study of adult striped bass throughout MA from 1997–2000, where crustaceans were found to represent ~45% of striped bass diet by weight within the North Shore region. While fish in our study consumed a slightly smaller proportion of crustaceans, the overall consumption of juvenile American lobster remained high.
Analysis of stable isotopes offers a more holistic examination of diet ontogeny as we could sample all fish (including fish with empty stomachs) and because isotopic signatures integrate across longer periods (Post, 2002). As indicated by stable carbon isotopic signatures, striped bass consumed organisms from both benthic and pelagic environments during the beginning of the spring/summer migration into MA. As the summer progressed, large striped bass bass relied heavily on benthic prey. Given the time lag between prey consumption and assimilation into muscle tissue (Buchheister and Latour, 2010), it is plausible that large striped bass feed primarily on pelagic food sources before their immediate arrival into MA, followed by a switch to benthic prey in MA where there is higher availability of crustaceans such as American lobsters (Thunberg, 2007).
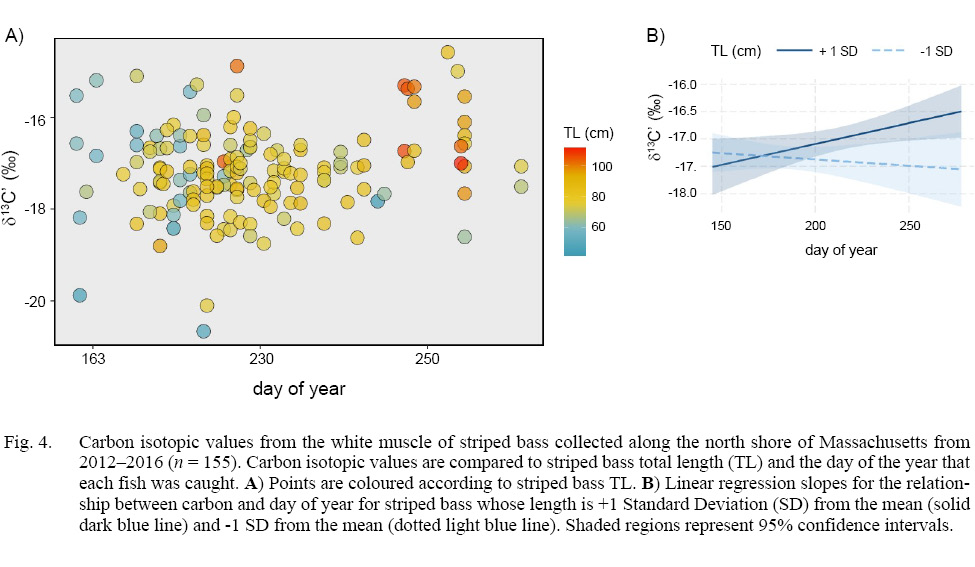
Fig. 4
This ontogenetic diet switch is somewhat counterintuitive given that fish prey offers more energy per gram wet weight (Steimle and Terranova, 1985) and since crustaceans, like the American lobster, are partly composed of chitin (Boßelmann et al., 2007), an organic material that is harder to digest than soft flesh. Analysis of striped bass LSI provides insight into possible explanations. Specifically, feeding on benthic organisms was slightly negatively correlated with the condition of small striped bass, as their livers weighed less relative to those of their pelagic-feeding counterparts. Conversely, benthic feeding seemed to significantly favour larger striped bass such that fish that fed on benthic prey items had larger livers, indicating that benthic feeding may allow these predators to build up better energy reserves. Given that striped bass experience decelerating growth by length, but their weight increases exponentially with age, large striped bass must propel a relatively heavier body through the w ater to capture prey. Chasing after fast-moving forage fish is thus potentially associated with high attack and pursuit costs for larger striped bass, while smaller, more streamlined individuals may be more capable of efficiently searching for and capturing forage fish. This finding is supported by work from a lake ecosystem, where pelagic Eurasian perch, Perca fluviatilis, were more streamlined than Eurasian perch feeding in the littoral zone (Quevedo et al., 2009).
By consuming benthic decapod prey that are slower than forage fish and potentially easier to capture, large striped bass may be able to reduce the energetic costs associated with capturing prey; which aligns with OFT (Pyke et al., 1977). This feeding strategy would allow striped bass to acquire increased energy reserves, as suggested by our LSI analysis. Similarly, work by Sherwood et al. (2002) suggested that the burst speed required to capture prey is an important component of foraging activity costs. In a lake ecosystem, the authors measured the lactate dehydrogenase levels in the white muscle of yellow perch, Perca flavescens, which is a proxy for anaerobic metabolism and burst swimming activity. Predatory Yellow perch that exhibited ontogenetic variation in diet and shifted from consuming zooplankton to benthic invertebrates at first and later to large prey fish were able to reduce their anaerobic activity costs in a step-wise fashion with each diet switch. By resetting their activity costs after each ontogenetic prey switch, these fish were able to maintain growth and prevent a bioenergetic bottleneck. It is plausible that large striped bass switch to feeding more heavily on American lobsters and other large crustacean prey to reduce the metabolic costs of foraging. However, our findings do not rule out other explanations for this ontogenetic transition toward benthic invertebrates. For example, it is possible that smaller striped bass would benefit from consuming benthic invertebrates but are unable to due to morphological challenges (e.g., limited gape size) or resource competition from larger individuals.

Fig. 5
Collectively, our study illustrates the significance of ontogenetic diet transitions for predatory fish, whereby the apparent transition to benthic decapods by large striped bass highlights that diversity in prey availability is important for maintaining predator condition with growth. We provide explanations for the energetic basis for this diet ontogeny in striped bass that would be supported by OFT. Specifically, stomach content and stable isotope analyses suggest that diet is driven partly by ontogenetic processes, such that large striped bass may benefit energetically from the consumption of large crustaceans over forage fish prey. A proposed mechanism for this ontogenetic shift from piscivory to benthivory follows that smaller, more streamlined striped bass likely benefit from the consumption of energetically rich forage fish. Conversely, large striped bass may suffer from increased attacking or searching costs associated with pelagic feeding and, as such, likely transition to benthic feeding, as suggested by enhancement in condition. Future experimental, comparative, and modeling studies should continue to unpack these mechanisms, providing additional insights into the complex relationships between predator and prey. Moreover, this study illustrates the variability of some predator-prey interactions over time but suggests that OFT can help to anticipate the consequences of fluctuating species abundances and size-frequency, spatial, and temporal distributions.
Acknowledgments
We would like to thank Kelsey Schultz, Joe Carracappa, Suzanne Kent, and Christopher Baillie for helping with our laboratory and fieldwork. Randy Sigler and the members of his fishing camp provided numerous striped bass samples, for which we are very grateful. Greg Veprek always went out of his way to bring us on his boat to collect samples and we are extremely grateful for his generosity. We also thank two blind reviewers whose feedback greatly improved our manuscript.
References
42nd Northeast Regional Stock Assessment Workshop. 2006. 42nd SAW assessment summary report (Ed.), In N. F. S. C. U.S. Dep. Commer.
Adams, S., and McLean, R. 1985. Estimation of largemouth bass, Micropterus salmoides Lacépède, growth using the liver somatic index and physiological variables*. Journal of Fish Biology, 26: 111–126. https://doi.org/10.1111/j.1095-8649.1985.tb04248.x
Atlantic States Marine Fisheries Commission. 2014. Addendum IV to Amendment 6 to the Atlantic Striped Bass Interstate Fishery Management Plan.
Atlantic States Marine Fisheries Commission. 2017. River herring stock assessment update volume II: state‐specific reports.
Bigelow, H. B., and Schroeder, W. C. 1953. Fishes of the Gulf of Maine. US Government Printing Office Washington, DC.
Boreman, J., Lewis, R. R., and Dadswell, M. 1987. Atlantic coastal migration of striped bass. In Common strategies of anadromous and catadromous fishes. American Fisheries Society, Symposium 1. 1: 331–339
Boßelmann, F., Romano, P., Fabritius, H., Raabe, D., and Epple, M. 2007. The composition of the exoskeleton of two crustacea: The American lobster Homarus americanus and the edible crab Cancer pagurus. Thermochimica Acta, 463: 65–68. https://doi.org/10.1016/j.tca.2007.07.018
Buchheister, A., and Latour, R. J. 2010. Turnover and fractionation of carbon and nitrogen stable isotopes in tissues of a migratory coastal predator, summer flounder (Paralichthys dentatus). Canadian Journal of Fisheries and Aquatic Sciences, 67: 445–461. https://doi.org/10.1139/F09-196
Carpenter, S. R., Kitchell, J. F., and Hodgson, J. R. 1985. Cascading trophic interactions and lake productivity. BioScience: 634–639. https://doi.org/10.2307/1309989
Chapoton, R. B., and Sykes, J. E. 1961. Atlantic coast migration of large striped bass as evidenced by fisheries and tagging. Transactions of the American Fisheries Society, 90:13–20. https://doi.org/10.1577/1548-8659(1961)90[13:ACMOLS]2.0.CO;2
Connell, J. H. 1961. Effects of competition, predation by Thais lapillus, and other factors on natural populations of the barnacle Balanus balanoides. Ecological Monographs 31: 61–104. https://doi.org/10.2307/1950746
Denno, R. F., and Lewis, D. 2009. Predator-prey interactions. The Princeton Guide to Ecology. Princeton University press, UK. https://doi.org/10.1515/9781400833023.202
Dovel, W. L. 1968. Predation by striped bass as a possible influence on population size of the Atlantic croaker. Transactions of the American Fisheries Society, 97: 313–319. https://doi.org/10.1577/1548-8659(1968)97[313:PBSBAA]2.0.CO;2
Dunning, D. J., Waldman, J. R., Ross, Q. E., and Mattson, M. T. 1997. Use of Atlantic tomcod and other prey by striped bass in the lower Hudson River estuary during winter. Transactions of the American Fisheries Society, 126: 857–861. https://doi.org/10.1577/1548-8659(1997)126%3C0857:UOATAO%3E2.3.CO;2
Einfalt, L. M., and Wahl, D. H. 1997. Prey selection by juvenile walleye as influenced by prey morphology and behavior. Canadian Journal of Fisheries and Aquatic Sciences, 54: 2618–2626. https://doi.org/10.1139/f97-172
Fry, B. 1988. Food web structure on Georges Bank from stable C, N, and S isotopic compositions. Limnology and Oceanography, 33: 1182–1190. https://doi.org/10.4319/lo.1988.33.5.1182
Gardinier, M. N., and Hoff, T. B. 1982. Diet of striped bass in the Hudson River Estuary [Morone saxatilis, New York]. New York Fish and Game Journal, 29: 152–165
Greene, K. E., Zimmerman, J. L., Laney, R. W., and Thomas-Blate, J. C. 2009. Atlantic coast diadromous fish habitat: a review of utilization, threats, recommendations for conservation, and research needs. Atlantic States Marine Fisheries Commission Habitat Management Series, 464, 276.
Griffin, J., and Margraf, F. 2003. The diet of Chesapeake Bay striped bass in the late 1950s. Fisheries Management and Ecology, 10: 323–328. https://doi.org/10.1046/j.1365-2400.2003.00367.x
Hambright, K. D. 1991. Experimental analysis of prey selection by largemouth bass: role of predator mouth width and prey body depth. Transactions of the American Fisheries Society, 120: 500–508. https://doi.org/10.1577/1548-8659(1991)120%3C0500:EAOPSB%3E2.3.CO;2
Hawlena, D., and Schmitz, O. J. 2010. Herbivore physiological response to predation risk and implications for ecosystem nutrient dynamics. Proceedings of the National Academy of Sciences, 107: 15503–15507. https://doi.org/10.1073/pnas.1009300107
Hilborn, R., R. Amoroso, O., Bogazzi, E., Jensen, O. P., Parma, A. M., Szuwalski, C., and Walters, C. J. 2017. When does fishing forage species affect their predators? Fisheries Research, 191: 211–221. https://doi.org/10.1016/j.fishres.2017.01.008
Hill, J., Evans, J. W., and Van Den Avyle, M. J. 1989. Species Profiles: Life Histories and Environmental Requirements of Coastal Fishes and Invertebrates (South Atlantic). Striped bass. DTIC Document. https://doi.org/10.21236/ADA226928
Hoque, M., Yusoff, F., Law, T., and Syed, M. 1998. Effect of hydrogen sulphide on liver somatic index and Fulton’s condition factor in Mystus nemurus. Journal of Fish Biology, 52: 23–30. https://doi.org/10.1111/j.1095-8649.1998.tb01549.x
Hughes, R. N. 1990. Behavioural mechanisms of food selection. Springer-Verlag GmbH & Co. KG. https://doi.org/10.1007/978-3-642-75118-9
Juanes, F., Stouder, D., and Feller, K. 1994. What determines prey size selectivity in piscivorous fishes. In Stouder D. J, Fresh, K. L., and Feller R. J. (Eds.), Theory and application in fish feeding ecology. pp. 79–100, Carolina University Press, Columbia.
Kiljunen, M., Grey, J. Sinisalo, T., Harrod, C., Immonen, H., and Jones, R. I. 2006. A revised model for lipid‐normalizing δ13C values from aquatic organisms, with implications for isotope mixing models. Journal of Applied Ecology, 43: 1213–1222. https://doi.org/10.1111/j.1365-2664.2006.01224.x
Langton, R. W., and Center, N. F. 1982. A review of methods used for estimating gut evacuation rates and calculating daily ration for fish. Ref. Doc:77–07.
Le Cren, E. 1951. The length-weight relationship and seasonal cycle in gonad weight and condition in the perch (Perca fluviatilis). The Journal of Animal Ecology, 2:2: 201–219. https://doi.org/10.2307/1540
Manooch, C. S. 1973. Food habits of yearling and adult striped bass, Morone saxatilis (Walbaum), from Albemarle Sound, North Carolina. Chesapeake Science, 14: 73–86. https://doi.org/10.2307/1350872
Massachusetts Division of Marine Fisheries. 2016. Annual Report 2016. Department of Fish and Game. https://www.mass.gov/files/documents/2017/08/30/2016-dmf-annual-report.pdf
Milinski, M. 1982. Optimal foraging: the influence of intraspecific competition on diet selection. Behavioral Ecology and Sociobiology 11:109-115. https://doi.org/10.1007/BF00300099
Nelson, G. 2015. Massachusetts Striped Bass Monitoring Report for 2014. Massachusetts Division of Marine Fisheries TR-62.
Nelson, G. A., Chase, B. C., and Stockwell, J. 2003. Food habits of striped bass (Morone saxatilis) in coastal waters of Massachusetts. Journal of Northwest Atlantic Fishery Science, 32: 1–25. https://doi.org/10.2960/J.v32.a1
Nelson, G. A., Chase, B. C., and Stockwell, J. 2006. Population consumption of fish and invertebrate prey by striped bass (Morone saxatilis) from coastal waters of northern Massachusetts, USA. Journal of Northwest Atlantic Fishery Science, 36: 111–126. https://doi.org/10.2960/J.v36.m576
North Pacific Fishery Management Council. 2019. Bering Sea Fishery Ecosystem Plan.
Overton, A. S., Margraf, F. J., and May, E. B. 2009. Spatial and temporal patterns in the diet of striped bass in Chesapeake Bay. Transactions of the American Fisheries Society, 138: 915–926. https://doi.org/10.1577/T07-261.1
Post, D. M. 2002. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology, 83:703–718. https://doi.org/10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2
Post, D. M., Layman, C. A., Arrington, D. A., Takimoto, G., Quattrochi, J., and Montana, C. G. 2007. Getting to the fat of the matter: models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia, 152: 179–189. https://doi.org/10.1007/s00442-006-0630-x
Pyke, G. H., Pulliam, H. R. and Charnov, E. L. 1977. Optimal foraging: a selective review of theory and tests. Quarterly Review of Biology, 52: 137–154. https://doi.org/10.1086/409852
Quevedo, M., Svanbäck, R., and Eklöv, P.. 2009. Intrapopulation niche partitioning in a generalist predator limits food web connectivity. Ecology, 90: 2263–2274. https://doi.org/10.1890/07-1580.1
R Core Team. 2020. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Austria, 2015. ISBN 3-900051-07-0: URL http://www.R-project.org.
Sherwood, G. D., Pazzia, I., Moeser, A. Hontela, A., and Rasmussen, J. B. 2002. Shifting gears: enzymatic evidence for the energetic advantage of switching diet in wild-living fish. Canadian Journal of Fisheries and Aquatic Sciences, 59: 229–241. https://doi.org/10.1139/f02-001
Sherwood, G. D., Rideout, R. M., Fudge, S. B., and Rose, G. A. 2007. Influence of diet on growth, condition and reproductive capacity in Newfoundland and Labrador cod (Gadus morhua): insights from stable carbon isotopes (δ13C). Deep Sea Research Part II: Topical Studies in Oceanography, 54: 2794–2809. https://doi.org/10.1016/j.dsr2.2007.08.007
Skinner, M. M., Martin, A. A., and Moore, B. C. 2016. Is lipid correction necessary in the stable isotope analysis of fish tissues? Rapid Communications in Mass Spectrometry, 30: 881–889. https://doi.org/10.1002/rcm.7480
Steimle, F., and Terranova, R. J. 1985. Energy equivalents of marine organisms from the continental shelf of the temperate Northwest Atlantic. Journal of Northwest Atlantic Fishery Science, 6: 117–124. https://doi.org/10.2960/J.v6.a11
Thunberg, E. M. 2007. Demographic and economic trends in the northeastern United States lobster (Homarus americanus) fishery, 1970–2005. US Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service, Northeast Fisheries Science Center.
Townsend, C. R., and Winfield, I. J. 1985. The application of optimal foraging theory to feeding behaviour in fish. Fish energetics. Springer, pp. 67–98. https://doi.org/10.1007/978-94-011-7918-8_3
Trussell, G. C., Ewanchuk, P. J., and Matassa, C. M. 2006. The fear of being eaten reduces energy transfer in a simple food chain. Ecology, 87: 2979–2984.
Zale, A. V., Parrish, D. L. and Sutton, T. M. 2012. Fisheries techniques. American Fisheries Society.
Zanden, M., and Rasmussen, J. B. 2001. Variation in δ15N and δ13C trophic fractionation: implications for aquatic food web studies. Limnology and Oceanography, 46: 2061–2066. https://doi.org/10.4319/lo.2001.46.8.2061
Citation: Murphy Jr., R.D., Nelson, G.A., and Grabowski, J.H. 2022. The feeding ecology of striped bass and the role of ontogeny. J. Northw. Atl. Fish. Sci., 53: 35–45. https://doi.org/10.2960/J.v53.m737